Building a blood-brain barrier penetration model in Flare™
Giovanna Tedesco†, Ryuchiro Hara†
†Cresset, New Cambridge House, Bassingbourn Road, Litlington, Cambridgeshire, SG8 0SS, UK
In this blog, we used the enhanced QSAR modeling capabilities in Flare V5 (due for release soon) to build a blood-brain barrier (BBB) permeation classification model using molecular descriptors imported from the RDKit by means of a Flare Python extension written ad hoc.
Introduction
Forge, our ligand-based design workbench, provides Field QSAR and Machine Learning methods that use descriptors based on electrostatic molecular fields and steric properties to characterize each molecule and build 3D-QSAR models of biological activity. These descriptors are very efficient at modeling specific ligand-protein interactions and derive quantitative models that relate the chemical structure of the ligand to the biological activity at the target of interest.
However, when building QSAR models for ADMET properties not involving a direct ligand-protein interaction, but rather a passive diffusion mechanism as for example BBB permeation, whole molecule properties such as Molecular Weight (MW), Polar Surface Area (PSA) and lipophilicity (i.e., SlogP) may be more appropriate. Indeed, many practitioners will want to extend these further to build QSAR models using calculated, experimental or custom properties. Although this is not possible in Forge, this will be possible in Flare V5. This upcoming release will incorporate all the functionality of Forge and make it available through Flare’s Python API to enable more complex QSAR workflows as demonstrated.
The data set
The data set used in this study is taken from Roy et al., ACS Omega 2019. In this paper, BBB permeation data was collected from different sources to generate a set of 1866 compounds, of which 1438 were classified as BBB permeating (BBB+, Class 1), and 428 as non-BBB permeating (BBB-, Class 0). The authors split the initial dataset into a training set of 1400 molecules and a test set of 466 compounds.
The compounds were imported in Flare, generating a training set of 1376 compounds (1058 BBB+/Class 1 and 318 BBB-/Class 0) and a test set of 459 compounds (354 BBB+/Class 1 and 105 BBB-/Class 0). Some molecules with invalid SMILES were not imported.
The initial model
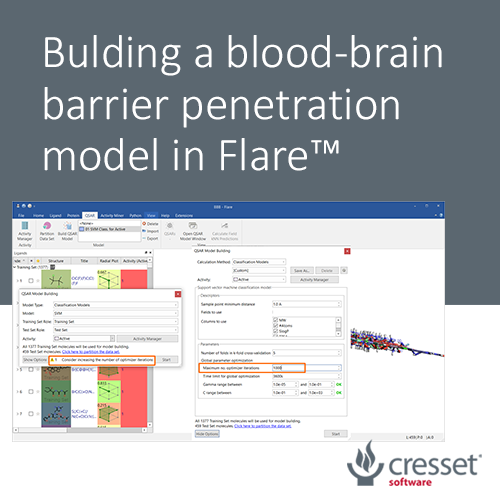
An initial Support Vector Machine (SVM) classification model was built using only Flare built-in descriptors, namely MW, SlogP, Topological Polar Surface Area (TPSA), Flexibility, number of rotatable bonds and total charge. As the data set is large, the maximum number of optimizer iterations was increased to 1000, as suggested by the strategy warning in the QSAR Model Building widget (Figure 1). All other options were used with their default values.
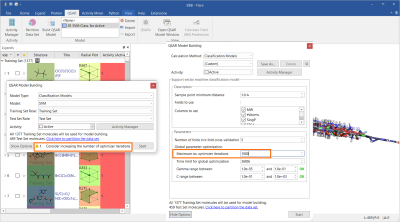
Figure 1. The initial SVM classification model was built using six Flare built-in descriptors.
The performance of this initial model is summarized by the confusion matrix (Figure 2 – left). The confusion matrix shows prediction frequencies for the actual classes for the training data vs. the classes predicted by the model. If all the off-diagonal values are zero, then all of the model predictions are correct. Other useful statistics derived from this matrix are reported for each role:
- Precision: The proportion of predictions that are actually correct, i.e., when the model predicts ‘Class 1’, how often it is correct? This value ranges from 0 (the model never makes a correct prediction) to 1 for perfect predictions.
- Recall: The proportion of molecules in a certain class that have been correctly identified as such, i.e., the number of correct Class 1 predictions divided by the total number of Class 1 molecules. This value ranges from 0 (the model never makes a correct prediction), to 1 for perfect predictions.
- Informedness: The probability of an informed decision being made rather than a random guess, ranging from 0 when your model is as good as a random guess, to 1 for perfect predictions.
The statistics reported in the confusion matrix are the average of the individual statistics for each class, which you can review in the project log (Figure 2 - right).
For machine learning models, the statistics for the training set/training set CV tend to be over-optimistic. The real predictive ability of the model is given by its performance on the test set, a set of molecules which is not included in the building of the model.
With a Precision of 0.87, a Recall of 0.83, an Informedness of 0.66 on the test set (Figure 2 - left), this very simple model already has a reasonably good ability to correctly classify the compounds in the test set as BBB+ (Class 1) vs. BBB- (Class 0), as shown in Figure 2. This is not totally surprising as we included descriptors such as MW, SlogP, TPSA and total charge which are known to be relevant in modeling passive diffusion processes.
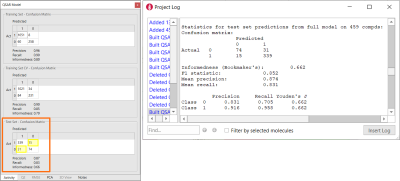
Figure 2. Left: Confusion matrix showing the classification performance of the BBB permeation model built on Flare descriptors. The yellow boxes mark the number of compounds incorrectly classified by the model. Right: the details of the QSAR model are reported in the Flare project log.
Importing custom descriptors in Flare
Although lipophilicity, molecular weight and polar surface area are important factors in determining BBB permeation, it is well known in the literature that other properties may contribute, for example the total number of H-bond donor and acceptors. More effective modeling of ADMET properties may also require the use of custom properties, such as for example calculated or experimental pKa or LogP/LogD, HOMO, LUMO and desolvation properties.
Naturally, these descriptors can be calculated separately and imported in Flare, but this is a manual and tedious process. The Flare Python API makes it easy to automate this type of workflow and create Flare ‘extensions’ which interface Flare to your favourite calculator of physico-chemical properties, or which pull experimental data from corporate databases.
For this study, the ‘Generate features for machine learning’ extension to Flare (Figure 3) was created to import descriptors for each compound from the RDKit and other sources, adding them as additional columns in the Flare project.
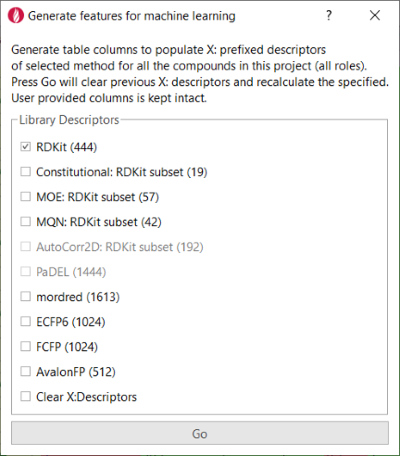
Figure 3. The Flare Python API makes it easy to write custom Flare Python extensions.
Building a refined BBB permeation model
After importing the descriptors from the RDKit, a new SVM model was built using the following descriptors:
MW, SlogP, TPSA, Flexibility, #RB, Total Formal Charge, NumHAcceptors, NumHDonors, RingCount
The first 6 (MW, SlogP, TPSA, Flexibility, #RB, Total Formal Charge) are calculated by Flare; NumHAcceptors, NumHDonors, RingCount by the RDKit.
The maximum number of optimizer iterations was increased to 1,000, and all the other settings were kept in their default values. The model obtained (Figure 4) shows a slightly improved performance.
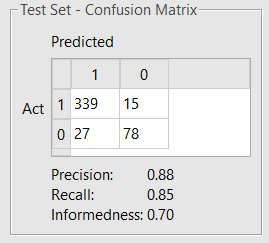
Figure 4. Confusion matrix showing test set classification performance of the refined BBB permeation model built on a mix of Flare and RDKit descriptors.
A model using all 444 RDKit descriptors shows a further slight improvement on the statistics of the test set (Precision 0.91, Recall 0.86, Informedness 0.73), at the cost of a much higher complexity and higher risk of overtraining the model.
Conclusion
Forge QSAR methods will be fully integrated into the next release of Flare V5 due for release summer 2021, featuring a new capability to work on custom descriptors. The Flare Python API enables you to easily create extensions connecting Flare to your favourite physico-chemical property calculator, or pulling data from your corporate databases. Together, these two features make it easy to build predictive QSAR models for ADMET properties such as BBB permeability for your own datasets.
Contact us to find out more about the integration of Flare into Forge, or request an evaluation.





















