Building QSAR models with RDKit Fingerprints and 2D descriptors in Flare™

Within Flare, the comprehensive software platform for ligand-based and structure-based drug design, users can build 3D Quantitative Structure-Activity Relationship (QSAR) models using 3D descriptors of electrostatic and shape. These models assume that the ligands in the dataset share a common binding mode with the active site of the biological target of interest.
However, in many cases, designers may wish to build a QSAR model for those biological properties where there is not a specified binding pose or event. For example, they may want to model an ADMET endpoint using a diverse set of ligands where there may not be an active site binding event, or this is not consistent for all the ligands. In such circumstances, alternative physico-chemical descriptors of the ligands need to be used, for example 2D descriptors such as Molecular Weight (MW), logP, Topological Polar Surface Area (TPSA), and/or the number of rotatable bonds.
Recent updates to Flare have fully integrated the generation of RDKit 2D descriptors1 and added the ability to use RDKit fingerprints1 for QSAR model building. In a previous article we showcased how users can build a Blood-Brain Barrier (BBB) permeation QSAR model, using 2D descriptor imported into Flare with a Python script. In this feature, we will revisit this study, using the newly built-in functionality to quickly and effortlessly generate RDKit descriptors.
QSAR model type
The BBB dataset used in this study is taken from the work of Roy et al.,2 and is a classification dataset, where a continuous BBB permeation measurement is not given. The reported response is a two-category classification: molecules are either being able to ‘pass’ the BBB and given a value of 1, or ‘can’t pass’ and get a value of 0. As such, we will build a classification QSAR model using these classes.
Importing RDKit descriptors for QSAR model building
Firstly, the full dataset was split into a training set, consisting of 70% of the total dataset (1284 ligands), and a test set, containing the remaining 30% of the dataset (551 ligands). Following this, RDKit 2D descriptors were generated by clicking on the ‘Import RDKit descriptors button’ found in the ‘Column & Activity Editor’ (Figure 1). You can choose to import all the RDKit descriptors, by choosing ‘Select All’, or to only import the descriptors you deem necessary, by ticking the select boxes.
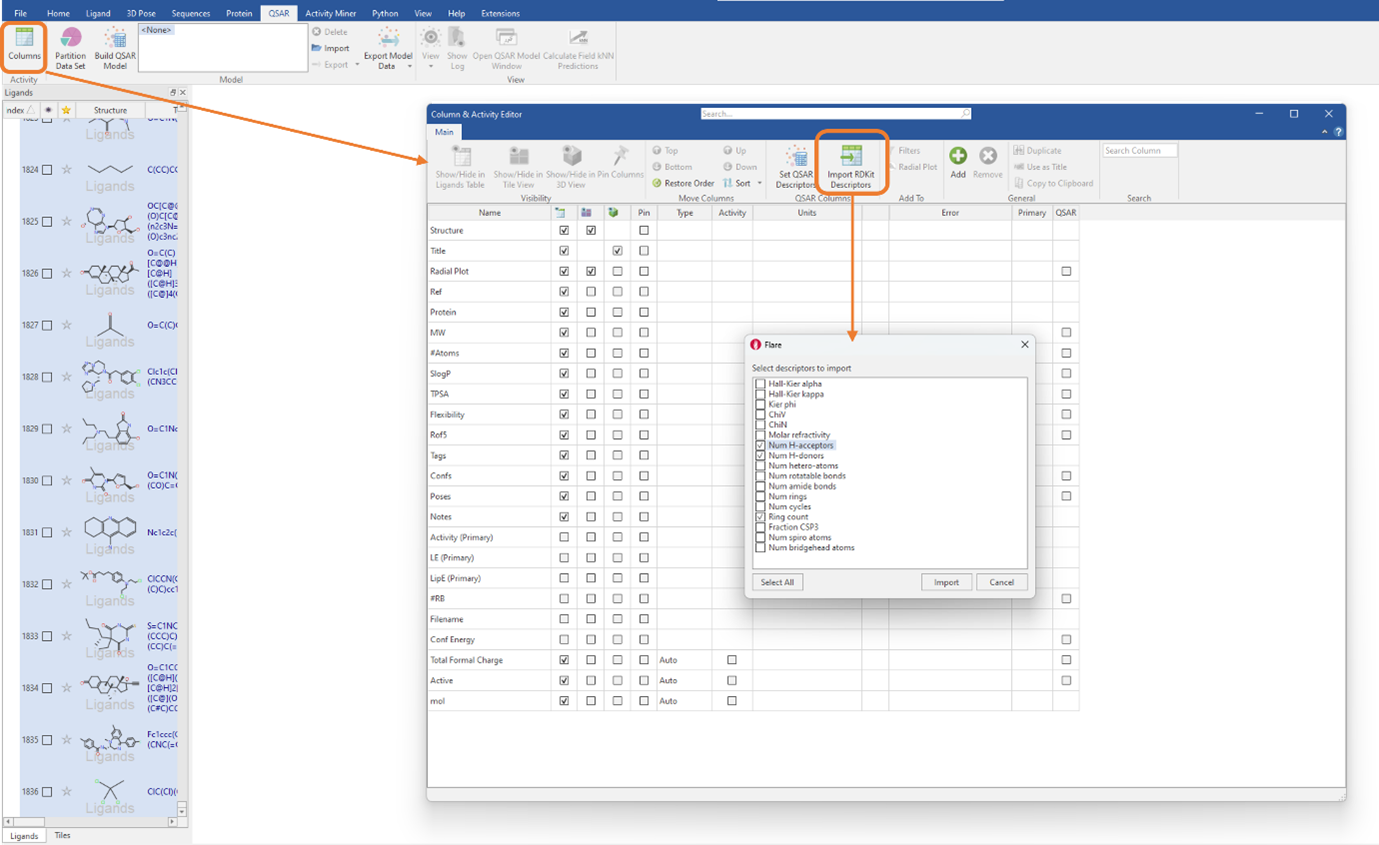
Figure 1: RDKit 2D physico-chemical descriptors can be imported by clicking a single button in the ‘Column & Activity Editor’.
Each RDKit descriptor you choose to import will be added as a molecular property column in the Ligands table. To use these descriptors for QSAR model building, we need to select the descriptors in the table then click the ‘Set QSAR Descriptors’ button in the Column & Activity Editor (Figure 2). For this experiment, we will be using the MW, SlogP, TPSA, Flexibility, #RB, Total Formal Charge, NumHAcceptors, NumHDonors and RingCount descriptors. The latter three descriptors are those previously imported from the RDKit (Figure 1).
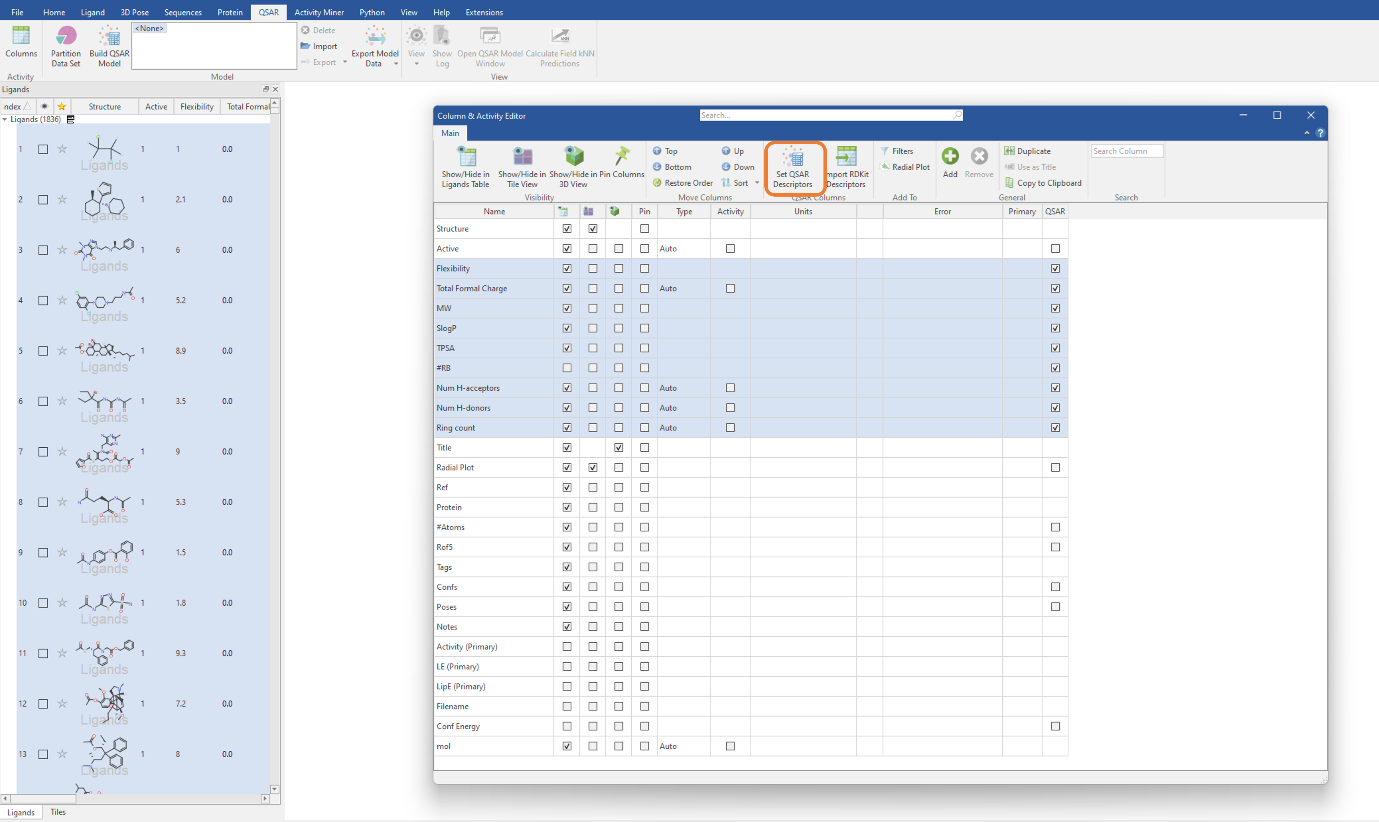
Figure 2: Selecting the imported RDKit descriptors to be used in our classification QSAR model.
To keep analogies to the previous study, we will build a SVM classification QSAR model using a combination of Flare generated and imported RDKit descriptors listed above (Figure 2). We need to ensure that the ‘Electrostatic’ and ‘Volume’ tick boxes are unticked. Additionally, it is worth noting here that as we are not using the electrostatic and shape field point 3D descriptors, our ligand dataset does not need to be aligned (Figure 3).
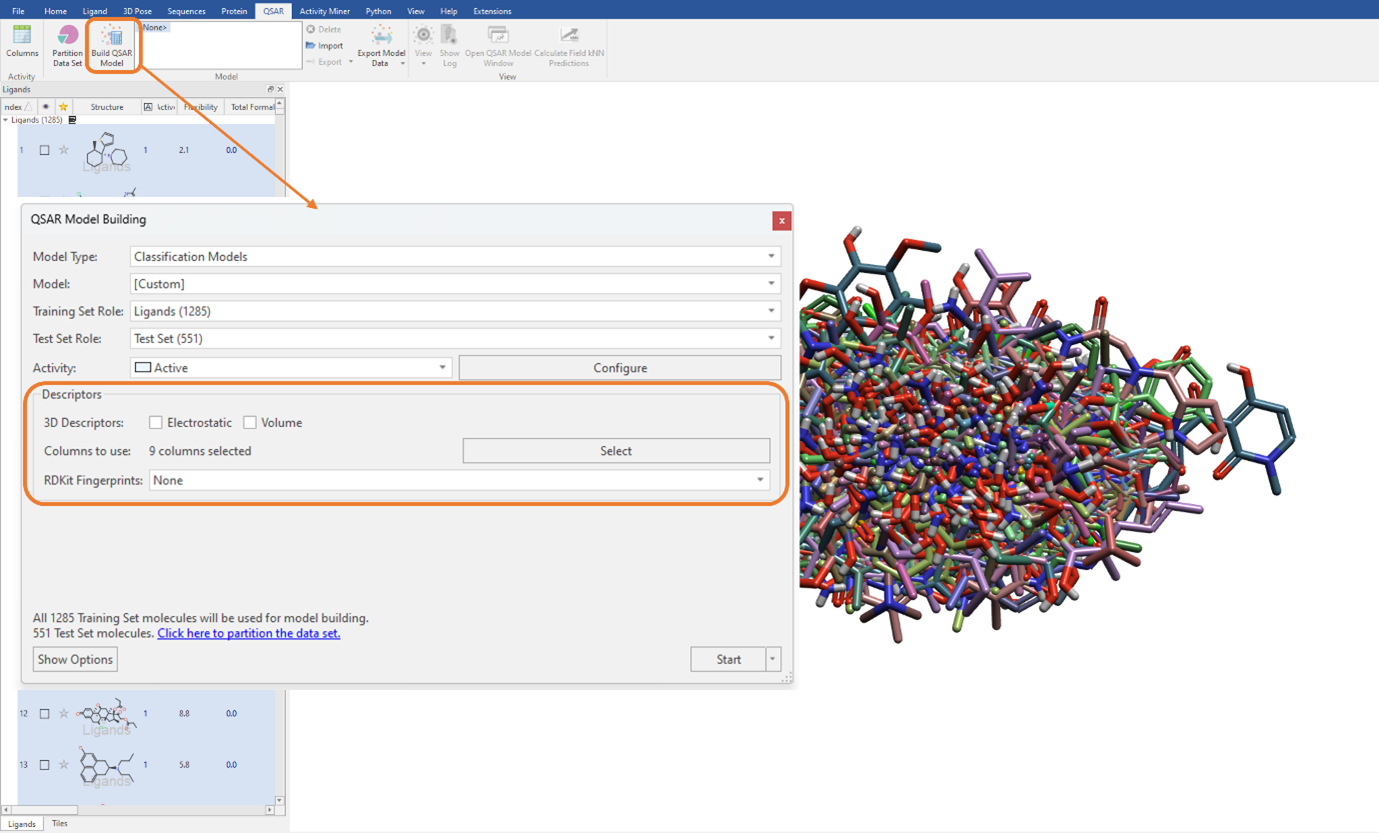
Figure 3: The SVM classification model was built using a combination of Flare generated and imported RDKit descriptors.
We can execute the QSAR model building calculation using an unlimited maximum number of optimizer iterations, which is the default for large datasets but can be manually edited in the ‘Show Options’ menu (Figure 3). The performance of the classification QSAR model, that being how well the classification model can correctly predict which ligand in the test set is assigned to category 0 or 1, is summarised by the Test Set Confusion Matrix (Figure 4). The off-diagonal elements show the frequency of incorrect predictions, namely a ligand belonging to category 0 being assigned to category 1 and vice versa. Off-diagonal values of zero indicate the classification QSAR model assigns each ligand in the test set to correct category 100% of the time.
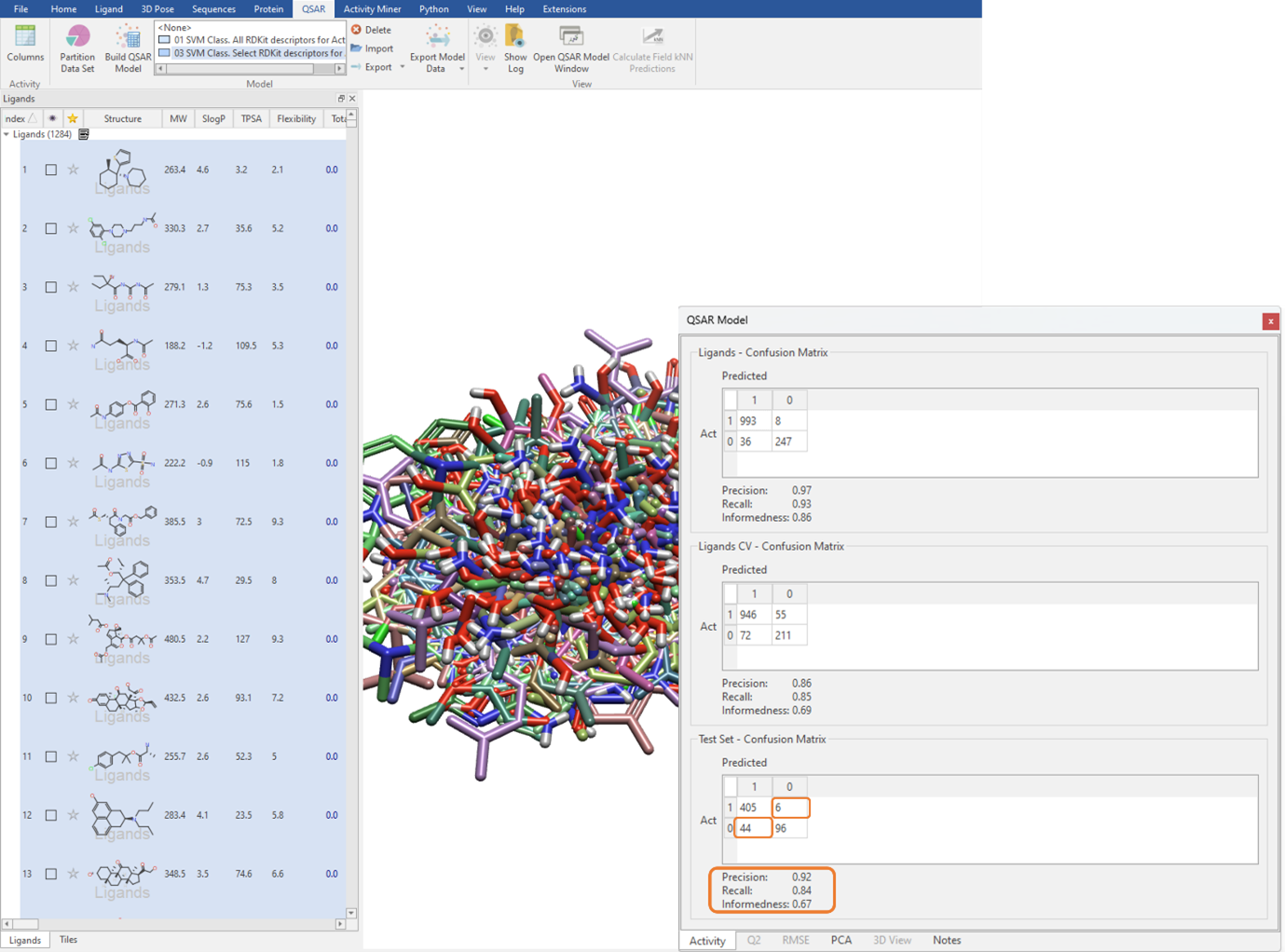
Figure 4: Using the Test Set Confusion Matrix to identify the predictive power of the classification QSAR model.
The Precision, Recall and Informedness are other statistical metrics derived from the Confusion Matrix (Figure 4). Precision describes the proportion of correct categorical assignments. Recall further granularizes this and reports the proportion of correct categorical assignments for a given category, essentially the number of correct category 1 predictions divided by the total number of category 1 ligands. Informedness describes the probability of an informed decision being made rather than a random guess. The closer all these values are to one, the more confidence we have in our classification QSAR model.
With a Precision of 0.92, a Recall of 0.84, and an Informedness of 0.67 on the test set (Figure 4), we can have statistical confidence in deploying this classification QSAR model to categorize novel ligands as either BBB permeable or impermeable. Furthermore, as it can be seen from the Test Set – Confusion Matrix, 6 ligands which are active are predicted to be inactive and 44 ligands which are inactive are predicted to be active.
These results suggest that, although we are predicting 44 false positives, we only have a small number (6) of false negatives. This means that, given that BBB penetration is a desirable property for our project, when it comes to prioritizing molecules for testing we have a low likelihood of missing a molecule that could potentially permeate the BBB and be of interest to the project.
Using RDKit fingerprints for QSAR model building
In Figure 3, you can see there is also the option to use RDKit fingerprints as a descriptor to build a QSAR model. RDKit fingerprints are a substructure fingerprint used to assess molecular similarity. By selecting this option, these fingerprint descriptors can be used in addition to, or instead of, the currently selected QSAR descriptors. Again, as with the 2D physico-chemical descriptors (Figure 1), these can be seamlessly used for model building by simply choosing the desired option from the ‘RDKit Fingerprints’ drop-down menu (Figure 5). The current choice includes ‘RDKit’ fingerprints, which we will use for this example, as well as Morgan fingerprints and MACCS keys.
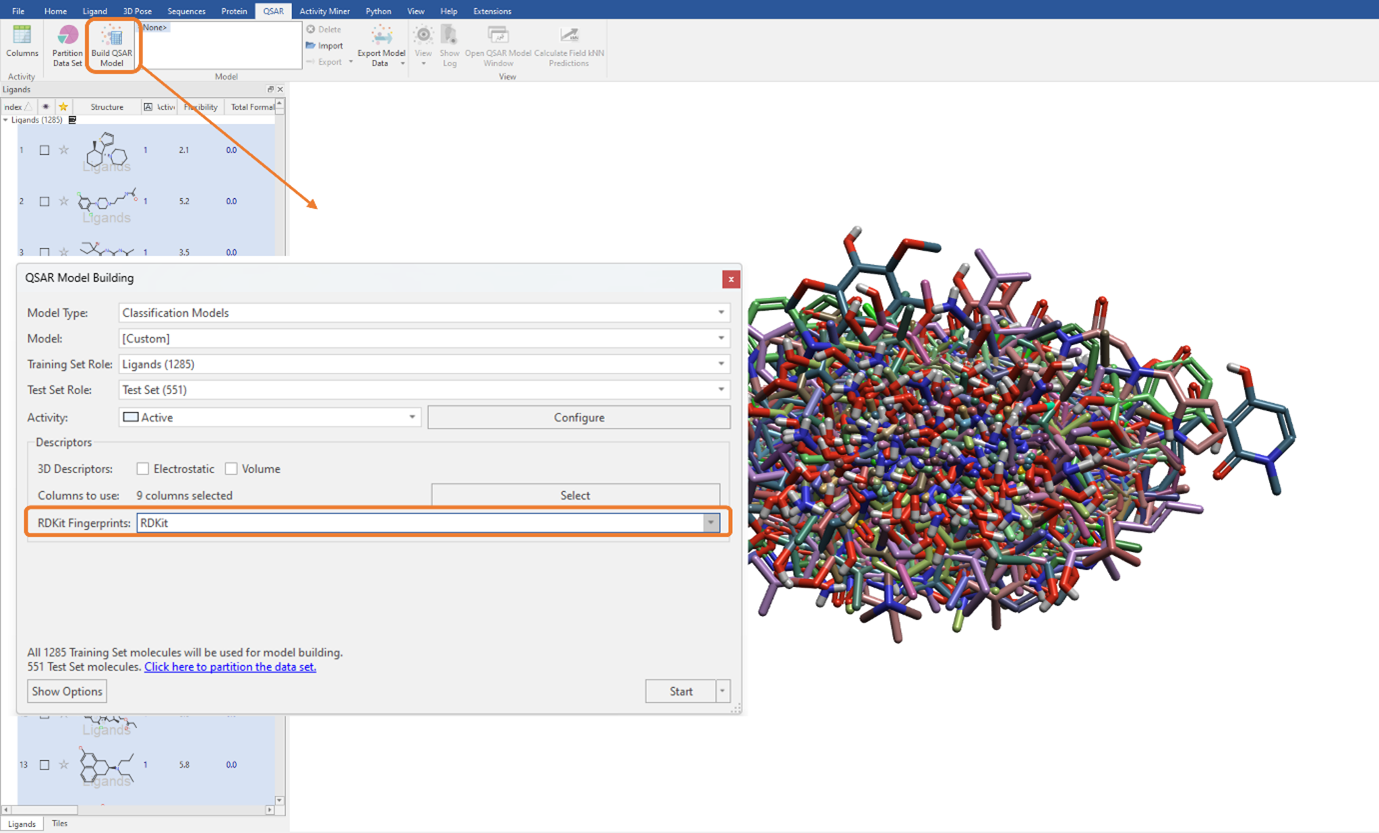
Figure 5: Using RDKit fingerprints to build the classification QSAR model. Note, we have de-selected the 3D and 2D descriptors (‘Columns to use’ is set to zero) and are only using the RDKit fingerprints for QSAR model building.
Upon building the classification QSAR model using the RDKit fingerprints, we obtain a Precision of 0.90, a Recall of 0.86, and an Informedness of 0.72, highlighting a slight improvement in the capability to predict BBB permeation compared to the classification QSAR model built using the RDKit 2D descriptors.
Conclusions
RDKit 2D descriptors and fingerprints can be quickly and easily imported and used for building QSAR models in Flare. In this example, these descriptors have been used to successfully to build classification QSAR models to predict the permeability of ligands across the BBB. Using RDKit descriptors or RDKit fingerprints offers an alternative to using Cresset 3D descriptors of electrostatic and shape for building predictive QSAR models of activity and ADMET properties.
Request a free evaluation of Flare today to further explore its full portfolio of molecular modelling capabilities. As part of the evaluation process, you’ll receive full support in installing the platform and accessing its wide range of features, while having the freedom to publish any results produced and use these for further research.
References
- Getting Started with the RDKit in Python — The RDKit 2023.03.1 documentation. https://www.rdkit.org/docs/GettingStartedInPython.html#list-of-available-descriptors (accessed 2023-07-31).
- Roy, D.; Hinge, V. K.; Kovalenko, A. To Pass or Not To Pass: Predicting the Blood–Brain Barrier Permeability with the 3D-RISM-KH Molecular Solvation Theory. ACS Omega 2019, 4 (16), 16774–16780. https://doi.org/10.1021/acsomega.9b01512.





















